6 Evaluate
The goal of syntheval is to make it simple to evaluate the utility and disclosure risk of synthetic data. It can be used with or without library(tidysynthesis).
6.1 Utility metrics
6.1.1 Setup
The following examples demonstrate utility and disclosure risk metrics using synthetic data based on the Palmer Penguins dataset (Horst, Hill, and Gorman 2020). library(syntheval) contains three built-in datasets:
penguins_conf: Pre-processedpenguinsdata that were passed into the synthesizer.penguins_postsynth: Apostsynthobject synthesized frompenguinsusinglibrary(tidysynthesis).penguins_syn_df: A data frame pulled frompenguins_postsynth. This is used to demonstrate howlibrary(syntheval)works with output from a synthesizer different thanlibrary(tidysynthesis).
Functions like util_proportions() and util_moments() have different behaviors for postsynth objects and data frames. By default, they only show synthesized variables for postsynth objects and show all common variables for data frames. The common_vars and synth_vars arguments can change this behavior.
6.1.2 Proportions
util_proportions() compares the proportions of classes from categircal variables in the original and synthetic data.
# A tibble: 2 × 5
# Groups: variable [1]
variable class synthetic original difference
<chr> <fct> <dbl> <dbl> <dbl>
1 sex female 0.529 0.495 0.0330
2 sex male 0.471 0.505 -0.0330All common variables are shown when using a data frame.
# A tibble: 8 × 5
# Groups: variable [3]
variable class synthetic original difference
<chr> <fct> <dbl> <dbl> <dbl>
1 island Biscoe 0.465 0.489 -0.0240
2 island Dream 0.414 0.369 0.0450
3 island Torgersen 0.120 0.141 -0.0210
4 sex female 0.529 0.495 0.0330
5 sex male 0.471 0.505 -0.0330
6 species Adelie 0.459 0.438 0.0210
7 species Chinstrap 0.234 0.204 0.0300
8 species Gentoo 0.306 0.357 -0.05116.1.3 Means and Totals
util_moments() compares the counts, means, standard deviations, skewness, and kurtoses of the original and synthetic data.
# A tibble: 20 × 6
variable statistic original synthetic difference proportion_difference
<fct> <fct> <dbl> <dbl> <dbl> <dbl>
1 bill_length_mm count 3.33e+2 333 0 0
2 bill_length_mm mean 4.40e+1 43.5 -0.502 -0.0114
3 bill_length_mm sd 5.47e+0 5.54 0.0723 0.0132
4 bill_length_mm skewness 4.51e-2 0.0646 0.0195 0.432
5 bill_length_mm kurtosis -8.88e-1 -0.948 -0.0598 0.0674
6 bill_depth_mm count 3.33e+2 333 0 0
7 bill_depth_mm mean 1.72e+1 17.3 0.122 0.00712
8 bill_depth_mm sd 1.97e+0 1.89 -0.0762 -0.0387
9 bill_depth_mm skewness -1.49e-1 -0.278 -0.129 0.867
10 bill_depth_mm kurtosis -8.97e-1 -0.742 0.155 -0.172
11 flipper_length… count 3.33e+2 333 0 0
12 flipper_length… mean 2.01e+2 199. -1.70 -0.00847
13 flipper_length… sd 1.40e+1 13.9 -0.135 -0.00961
14 flipper_length… skewness 3.59e-1 0.611 0.253 0.705
15 flipper_length… kurtosis -9.65e-1 -0.704 0.261 -0.270
16 body_mass_g count 3.33e+2 333 0 0
17 body_mass_g mean 4.21e+3 4162. -45.0 -0.0107
18 body_mass_g sd 8.05e+2 783. -22.3 -0.0277
19 body_mass_g skewness 4.70e-1 0.655 0.185 0.394
20 body_mass_g kurtosis -7.40e-1 -0.388 0.353 -0.477 util_totals() is similar to util_moments() but looks at counts and totals.
# A tibble: 8 × 6
variable statistic original synthetic difference proportion_difference
<fct> <fct> <dbl> <dbl> <dbl> <dbl>
1 bill_length_mm count 333 333 0 0
2 bill_length_mm total 14650. 14483. -167. -0.0114
3 bill_depth_mm count 333 333 0 0
4 bill_depth_mm total 5716. 5757. 40.7 0.00712
5 flipper_length_… count 333 333 0 0
6 flipper_length_… total 66922 66355 -567 -0.00847
7 body_mass_g count 333 333 0 0
8 body_mass_g total 1400950 1385950 -15000 -0.0107 6.1.4 Percentiles
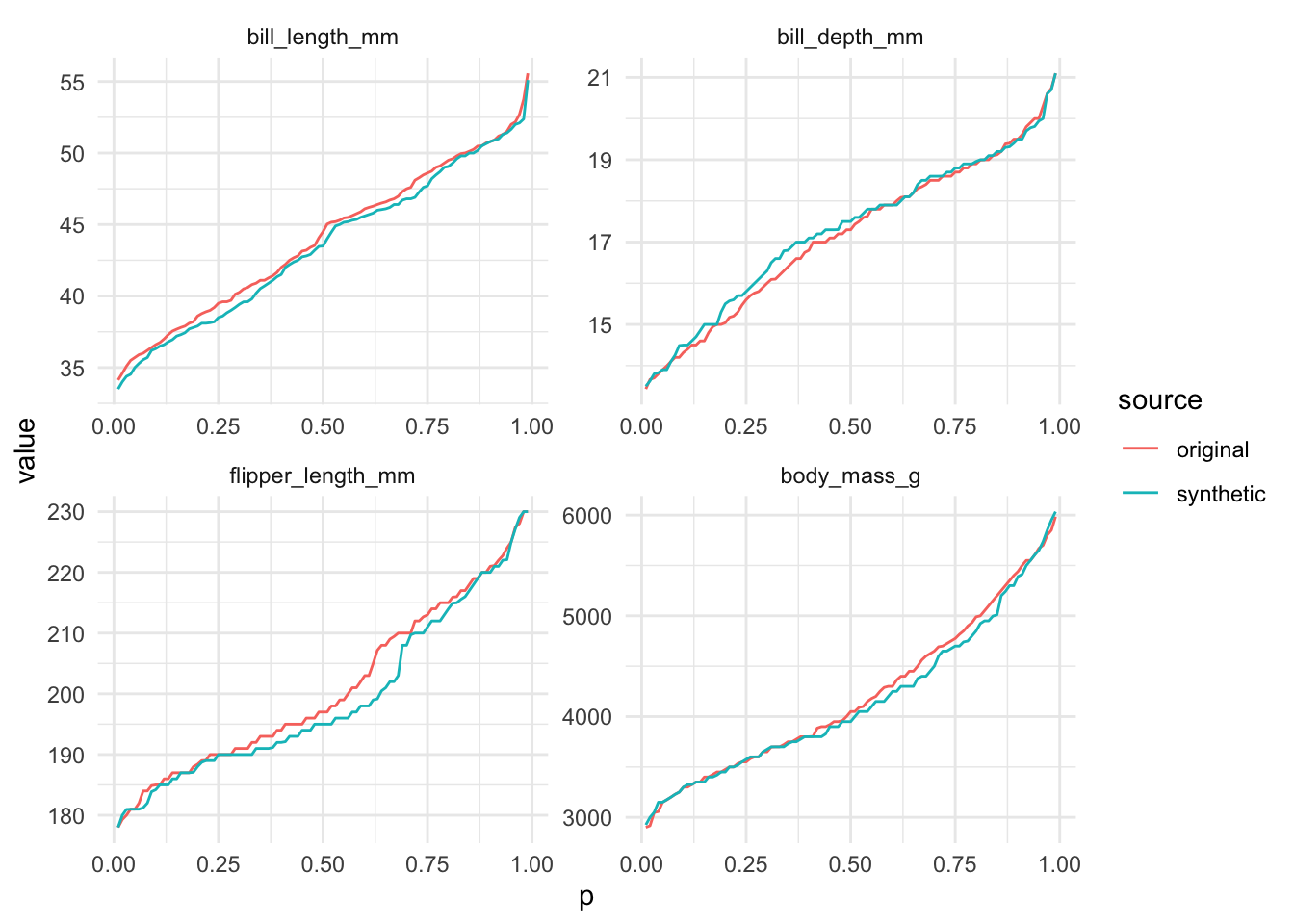
util_percentiles() compares percentiles from the original and synthetic data. The default percentiles are c(0.1, 0.5, 0.9) and can be easily overwritten.
# A tibble: 8 × 6
p variable original synthetic difference proportion_difference
<dbl> <fct> <dbl> <dbl> <dbl> <dbl>
1 0.5 bill_length_mm 44.5 43.5 -1 -0.0225
2 0.8 bill_length_mm 49.5 49.1 -0.440 -0.00889
3 0.5 bill_depth_mm 17.3 17.5 0.200 0.0116
4 0.8 bill_depth_mm 18.9 19.0 0.0600 0.00317
5 0.5 flipper_length_mm 197 195 -2 -0.0102
6 0.8 flipper_length_mm 215 214 -1 -0.00465
7 0.5 body_mass_g 4050 3950 -100 -0.0247
8 0.8 body_mass_g 4990 4850 -140 -0.0281 The functions are designed to work well with library(ggplot2).
6.1.5 KS Distance
util_ks_distance() shows the Kolmogorov-Smirnov distance between the original distribution and synthetic distribution for numeric variables. The function also returns the point(s) of the maximum distance.
# A tibble: 14 × 3
variable value D
<chr> <dbl> <dbl>
1 bill_length_mm 38.7 0.0601
2 bill_depth_mm 16.7 0.0511
3 bill_depth_mm 16.7 0.0511
4 bill_depth_mm 16.8 0.0511
5 bill_depth_mm 16.8 0.0511
6 flipper_length_mm 196. 0.0781
7 flipper_length_mm 196. 0.0781
8 flipper_length_mm 197. 0.0781
9 flipper_length_mm 197. 0.0781
10 flipper_length_mm 197. 0.0781
11 body_mass_g 4359. 0.0480
12 body_mass_g 4370. 0.0480
13 body_mass_g 4381. 0.0480
14 body_mass_g 4392. 0.04806.1.6 Co-Occurrence
util_co_occurence() differences the lower triangles of co-occurrence matrices calculated on numeric variables in the original data and synthetic data.
co_occurrence <- util_co_occurrence(
postsynth = penguins_postsynth,
data = penguins_conf
)
co_occurrence$co_occurrence_difference bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
bill_length_mm NA NA NA NA
bill_depth_mm 0 NA NA NA
flipper_length_mm 0 0 NA NA
body_mass_g 0 0 0 NAThe function returns the MAE for co-occurrences, which provides a sense of the median error between the original and synthetic data. The function also returns the RMSE for co-occurrences, which provides a sense of the average error between the original data and synthetic data.
All observations have non-zero bill_length_mm, bill_depth_mm, flipper_length_mm, and body_mass_g. util_co_occurrence() is most useful for economic variables like income and wealth where 0 is a common value.
6.1.7 Correlations
util_corr_fit() differences the lower triangles of correlation matrices calculated on numeric variables in the original data and synthetic data.
corr_fit <- util_corr_fit(
postsynth = penguins_postsynth,
data = penguins_conf
)
round(corr_fit$correlation_difference, digits = 3) bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
bill_length_mm NA NA NA NA
bill_depth_mm -0.069 NA NA NA
flipper_length_mm -0.003 -0.048 NA NA
body_mass_g 0.031 -0.011 -0.08 NAThe function returns the MAE for correlation coefficients, which provides a sense of the median error between the original and synthetic data. The function also returns the RMSE for the correlation coefficients, which provides a sense of the average error between the original synthetic data.
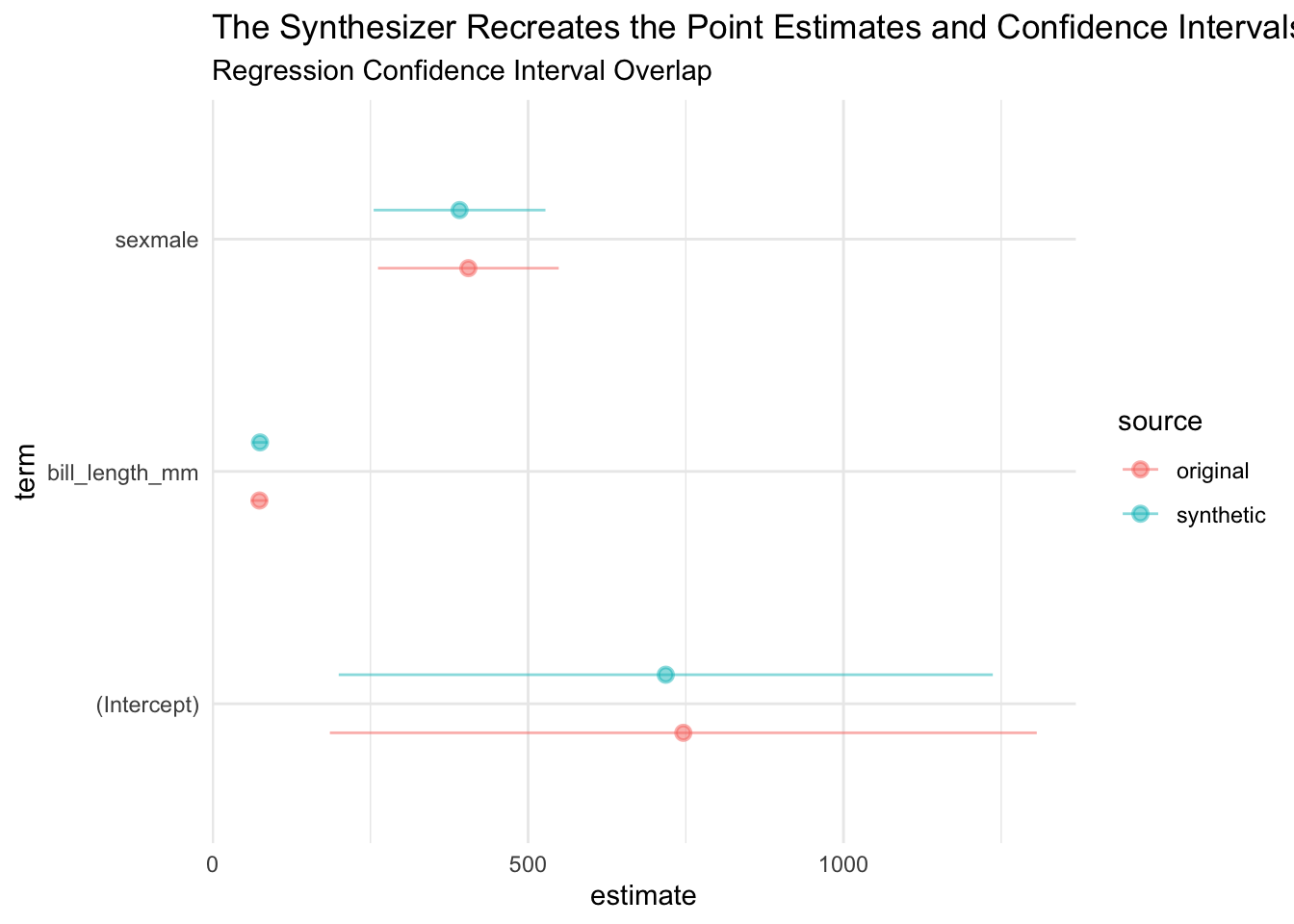
6.1.8 Confidence Interval Overlap
util_ci_overlap() compares a linear regression models estimated on the original data and synthetic data. formula specifies the functional form of the regression model.
$ci_overlap() summarizes each coefficient including how much the confidence intervals overlap, if the signs match, and if the statistical significance matches.
# A tibble: 3 × 8
term overlap coef_diff std_coef_diff sign_match significance_match ss_match
<chr> <dbl> <dbl> <dbl> <lgl> <lgl> <lgl>
1 (Inter… 0.963 -27.8 -0.0978 TRUE TRUE TRUE
2 bill_l… 0.965 0.917 0.138 TRUE TRUE TRUE
3 sexmale 0.951 -14.0 -0.192 TRUE TRUE TRUE
# ℹ 1 more variable: sso_match <lgl>$coefficient provides detail for each coefficient and is useful for data visualization.
ci_overlap$coefficient |>
ggplot(aes(x = estimate, xmin = conf.low, xmax = conf.high, y = term, color = source)) +
geom_pointrange(alpha = 0.5, position = position_dodge(width = 0.5)) +
labs(
title = "The Synthesizer Recreates the Point Estimates and Confidence Intervals",
subtitle = "Regression Confidence Interval Overlap"
)
6.1.9 Discriminant-Based Metrics
Discriminant-based metrics build models to predict if an observation is original or synthetic and then evaluate those model predictions. Ideally, it should be difficult for a model to distinguish, or discriminate, between original observations and synthetic observations.
- Any classification model that generates probabilities from
library(tidymodels)can be used to generate propensities (the estimated probability than observation is synthetic). - By default, the code creates a training/testing split and returns separate metrics for each split. This can be turned off with
split = FALSE. - The code can handle hyperparameter tuning.
- After calculating propensities, the code can calculate ROC AUC, SPECKS, pMSE, and pMSE ratio.
6.1.9.1 Example Using Decision Trees
Discriminant-based metrics are built a discrimination object created by discrimination().
Next, we use library(tidymodels) to specify a model. We recommend the tidymodels tutorial to learn more.
Next, we fit the model to the data to generate predicted probabilities.
At this point, we can use
add_discriminator_auc()to add the ROC AUC for the predicted probabilitiesadd_specks()to add SPECKS for the predicted probabilitiesadd_pmse()to add pMSE for the predicted probabilitiesadd_pmse_ratio(times = 25)to add the pMSE ratio using the pMSE model and 25 bootstrap samples
$combined_data
# A tibble: 666 × 8
.source_label species island sex bill_length_mm bill_depth_mm
<fct> <fct> <fct> <fct> <dbl> <dbl>
1 original Adelie Torgersen male 39.1 18.7
2 original Adelie Torgersen female 39.5 17.4
3 original Adelie Torgersen female 40.3 18
4 original Adelie Torgersen female 36.7 19.3
5 original Adelie Torgersen male 39.3 20.6
6 original Adelie Torgersen female 38.9 17.8
7 original Adelie Torgersen male 39.2 19.6
8 original Adelie Torgersen female 41.1 17.6
9 original Adelie Torgersen male 38.6 21.2
10 original Adelie Torgersen male 34.6 21.1
# ℹ 656 more rows
# ℹ 2 more variables: flipper_length_mm <dbl>, body_mass_g <dbl>
$propensities
# A tibble: 666 × 10
.pred_synthetic .source_label .sample species island sex bill_length_mm
<dbl> <fct> <chr> <fct> <fct> <fct> <dbl>
1 0.290 original testing Adelie Torgersen male 39.1
2 0.290 original training Adelie Torgersen fema… 39.5
3 0.290 original training Adelie Torgersen fema… 40.3
4 0.636 original testing Adelie Torgersen fema… 36.7
5 0.290 original training Adelie Torgersen male 39.3
6 0.290 original training Adelie Torgersen fema… 38.9
7 0.290 original training Adelie Torgersen male 39.2
8 0.290 original training Adelie Torgersen fema… 41.1
9 0.636 original testing Adelie Torgersen male 38.6
10 0.8 original training Adelie Torgersen male 34.6
# ℹ 656 more rows
# ℹ 3 more variables: bill_depth_mm <dbl>, flipper_length_mm <dbl>,
# body_mass_g <dbl>
$discriminator
══ Workflow [trained] ══════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: decision_tree()
── Preprocessor ────────────────────────────────────────────────────────────────
0 Recipe Steps
── Model ───────────────────────────────────────────────────────────────────────
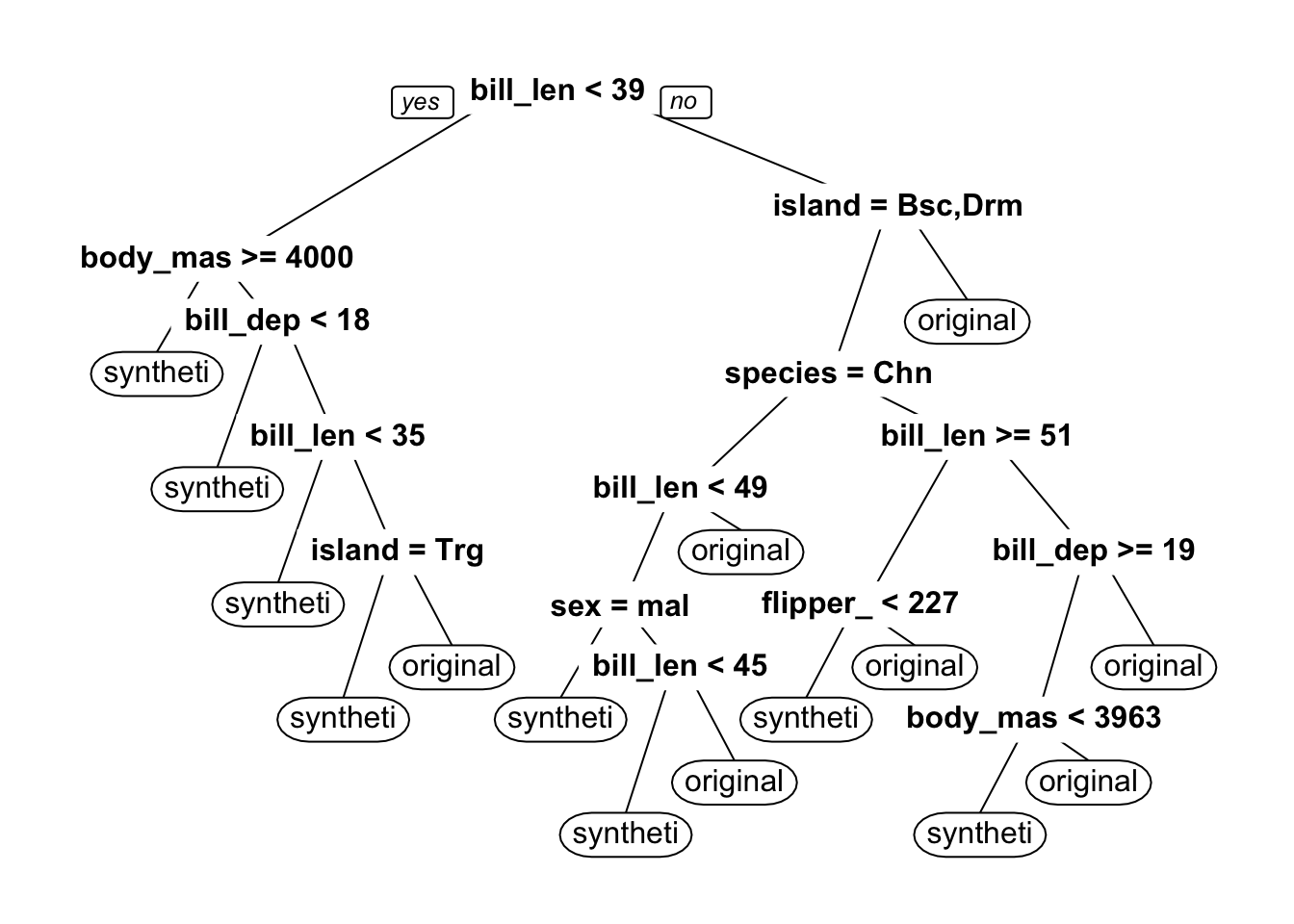
n= 498
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 498 249 synthetic (0.50000000 0.50000000)
2) bill_length_mm< 38.75 117 47 synthetic (0.59829060 0.40170940)
4) body_mass_g>=4000 15 3 synthetic (0.80000000 0.20000000) *
5) body_mass_g< 4000 102 44 synthetic (0.56862745 0.43137255)
10) bill_depth_mm< 17.85 62 21 synthetic (0.66129032 0.33870968) *
11) bill_depth_mm>=17.85 40 17 original (0.42500000 0.57500000)
22) bill_length_mm< 34.8 8 2 synthetic (0.75000000 0.25000000) *
23) bill_length_mm>=34.8 32 11 original (0.34375000 0.65625000)
46) island=Torgersen 11 4 synthetic (0.63636364 0.36363636) *
47) island=Biscoe,Dream 21 4 original (0.19047619 0.80952381) *
3) bill_length_mm>=38.75 381 179 original (0.46981627 0.53018373)
6) island=Biscoe,Dream 350 170 original (0.48571429 0.51428571)
12) species=Chinstrap 108 49 synthetic (0.54629630 0.45370370)
24) bill_length_mm< 49.15 66 23 synthetic (0.65151515 0.34848485)
48) sex=male 18 1 synthetic (0.94444444 0.05555556) *
49) sex=female 48 22 synthetic (0.54166667 0.45833333)
98) bill_length_mm< 45.35 22 6 synthetic (0.72727273 0.27272727) *
99) bill_length_mm>=45.35 26 10 original (0.38461538 0.61538462) *
25) bill_length_mm>=49.15 42 16 original (0.38095238 0.61904762) *
13) species=Adelie,Gentoo 242 111 original (0.45867769 0.54132231)
26) bill_length_mm>=51.2 27 10 synthetic (0.62962963 0.37037037)
52) flipper_length_mm< 226.5 16 3 synthetic (0.81250000 0.18750000) *
53) flipper_length_mm>=226.5 11 4 original (0.36363636 0.63636364) *
27) bill_length_mm< 51.2 215 94 original (0.43720930 0.56279070)
54) bill_depth_mm>=19.05 26 11 synthetic (0.57692308 0.42307692)
108) body_mass_g< 3962.5 13 2 synthetic (0.84615385 0.15384615) *
109) body_mass_g>=3962.5 13 4 original (0.30769231 0.69230769) *
55) bill_depth_mm< 19.05 189 79 original (0.41798942 0.58201058) *
7) island=Torgersen 31 9 original (0.29032258 0.70967742) *
$discriminator_auc
# A tibble: 2 × 4
.sample .metric .estimator .estimate
<fct> <chr> <chr> <dbl>
1 training roc_auc binary 0.700
2 testing roc_auc binary 0.475
$pmse
# A tibble: 2 × 4
.source .pmse .null_pmse .pmse_ratio
<fct> <dbl> <dbl> <dbl>
1 training 0.0356 0.0317 1.12
2 testing 0.0318 0.0320 0.995
$specks
# A tibble: 2 × 2
.source .specks
<fct> <dbl>
1 training 0.325
2 testing 0.0595
attr(,"class")
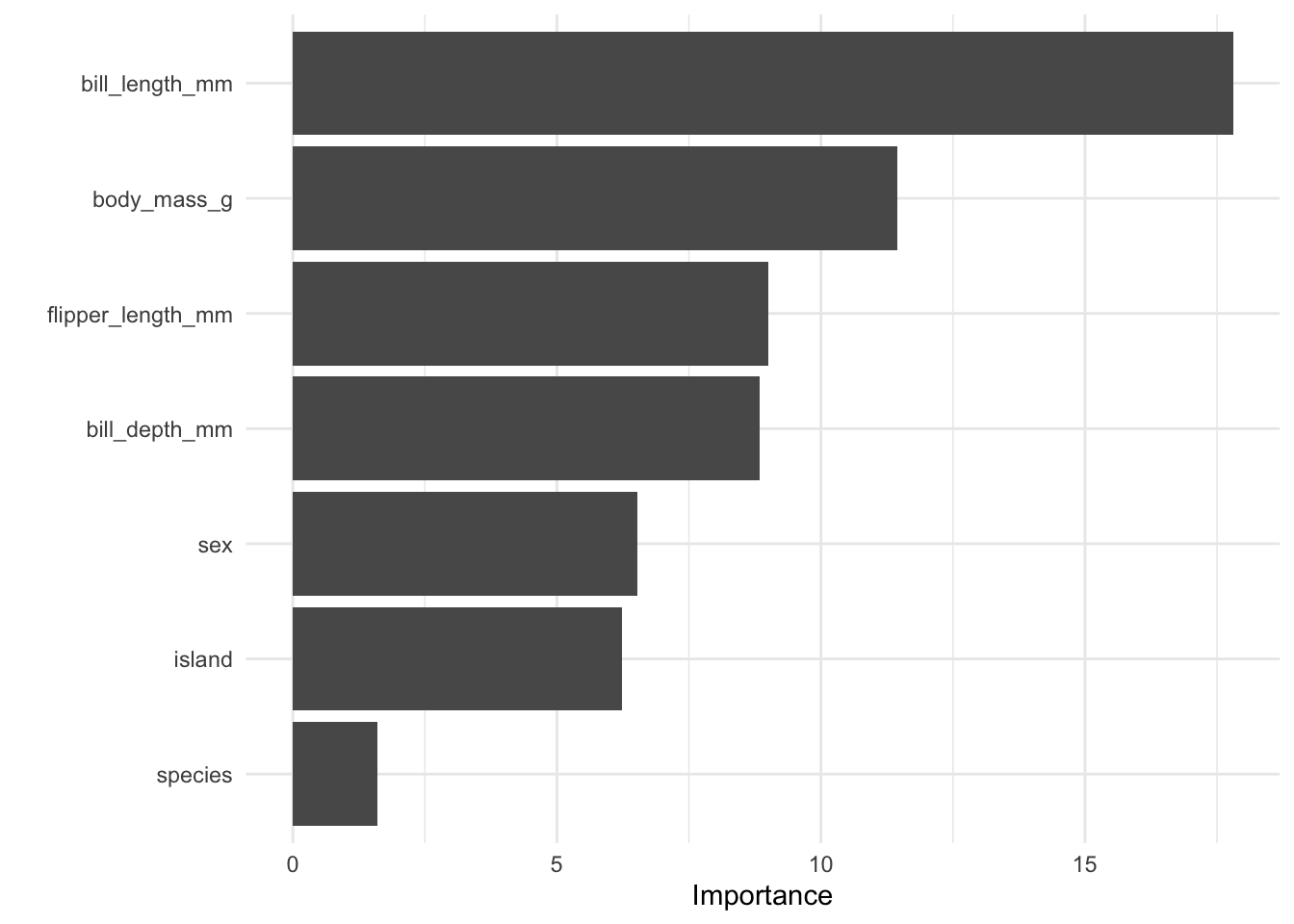
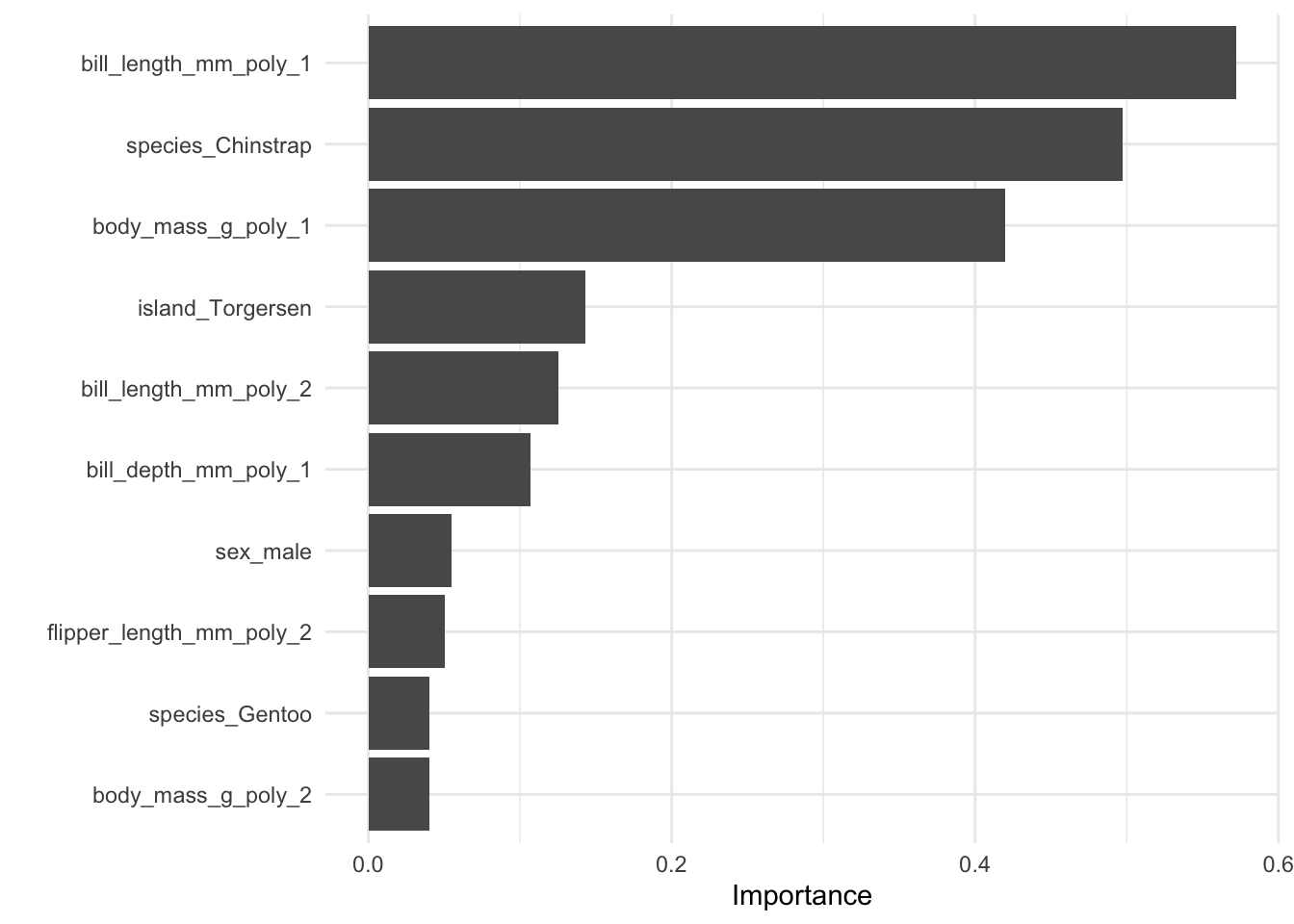
[1] "discrimination"Finally, we can look at variable importance and the decision tree from our discriminator.
6.1.9.2 Example Using Regularized Regression
Let’s repeat the workflow from above with LASSO logistic regression and hyperparameter tuning.
# create discrimination
disc2 <- discrimination(postsynth = penguins_postsynth, data = penguins_conf)
# create a recipe that includes 2nd-degree polynomials, dummy variables, and
# standardization
lasso_rec <- recipe(
.source_label ~ .,
data = disc2$combined_data
) %>%
step_poly(all_numeric_predictors(), degree = 2) %>%
step_dummy(all_nominal_predictors()) %>%
step_normalize(all_predictors())
# create the model
lasso_mod <- logistic_reg(
penalty = tune(),
mixture = 1
) %>%
set_engine(engine = "glmnet") %>%
set_mode(mode = "classification")
# create a tuning grid
lasso_grid <- grid_regular(penalty(), levels = 10)
# add the propensities
disc2 <- disc2 %>%
add_propensities_tuned(
recipe = lasso_rec,
spec = lasso_mod,
grid = lasso_grid
) # calculate metrics
disc2 %>%
add_discriminator_auc() %>%
add_specks() %>%
add_pmse() %>%
add_pmse_ratio(times = 25)$combined_data
# A tibble: 666 × 8
.source_label species island sex bill_length_mm bill_depth_mm
<fct> <fct> <fct> <fct> <dbl> <dbl>
1 original Adelie Torgersen male 39.1 18.7
2 original Adelie Torgersen female 39.5 17.4
3 original Adelie Torgersen female 40.3 18
4 original Adelie Torgersen female 36.7 19.3
5 original Adelie Torgersen male 39.3 20.6
6 original Adelie Torgersen female 38.9 17.8
7 original Adelie Torgersen male 39.2 19.6
8 original Adelie Torgersen female 41.1 17.6
9 original Adelie Torgersen male 38.6 21.2
10 original Adelie Torgersen male 34.6 21.1
# ℹ 656 more rows
# ℹ 2 more variables: flipper_length_mm <dbl>, body_mass_g <dbl>
$propensities
# A tibble: 666 × 10
.pred_synthetic .source_label .sample species island sex bill_length_mm
<dbl> <fct> <chr> <fct> <fct> <fct> <dbl>
1 0.410 original training Adelie Torgersen male 39.1
2 0.432 original training Adelie Torgersen fema… 39.5
3 0.325 original training Adelie Torgersen fema… 40.3
4 0.453 original training Adelie Torgersen fema… 36.7
5 0.340 original training Adelie Torgersen male 39.3
6 0.444 original testing Adelie Torgersen fema… 38.9
7 0.471 original training Adelie Torgersen male 39.2
8 0.332 original testing Adelie Torgersen fema… 41.1
9 0.366 original training Adelie Torgersen male 38.6
10 0.585 original testing Adelie Torgersen male 34.6
# ℹ 656 more rows
# ℹ 3 more variables: bill_depth_mm <dbl>, flipper_length_mm <dbl>,
# body_mass_g <dbl>
$discriminator
══ Workflow [trained] ══════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: logistic_reg()
── Preprocessor ────────────────────────────────────────────────────────────────
3 Recipe Steps
• step_poly()
• step_dummy()
• step_normalize()
── Model ───────────────────────────────────────────────────────────────────────
Call: glmnet::glmnet(x = maybe_matrix(x), y = y, family = "binomial", alpha = ~1)
Df %Dev Lambda
1 0 0.00 0.0309100
2 3 0.06 0.0281700
3 4 0.16 0.0256600
4 5 0.34 0.0233800
5 4 0.51 0.0213100
6 6 0.66 0.0194100
7 8 0.80 0.0176900
8 8 0.92 0.0161200
9 8 1.02 0.0146900
10 8 1.11 0.0133800
11 8 1.18 0.0121900
12 8 1.24 0.0111100
13 9 1.40 0.0101200
14 10 1.58 0.0092230
15 10 1.73 0.0084040
16 10 1.85 0.0076570
17 10 1.95 0.0069770
18 10 2.04 0.0063570
19 10 2.11 0.0057920
20 10 2.17 0.0052780
21 10 2.22 0.0048090
22 10 2.26 0.0043820
23 10 2.30 0.0039920
24 10 2.33 0.0036380
25 10 2.35 0.0033150
26 10 2.37 0.0030200
27 10 2.39 0.0027520
28 10 2.40 0.0025070
29 10 2.41 0.0022850
30 11 2.42 0.0020820
31 11 2.43 0.0018970
32 11 2.44 0.0017280
33 11 2.45 0.0015750
34 12 2.45 0.0014350
35 12 2.46 0.0013070
36 12 2.46 0.0011910
37 12 2.47 0.0010850
38 12 2.47 0.0009890
39 13 2.47 0.0009011
40 13 2.48 0.0008211
41 13 2.48 0.0007481
42 13 2.48 0.0006817
43 13 2.48 0.0006211
44 13 2.49 0.0005659
45 13 2.49 0.0005156
46 13 2.49 0.0004698
...
and 1 more lines.
$discriminator_auc
# A tibble: 2 × 4
.sample .metric .estimator .estimate
<fct> <chr> <chr> <dbl>
1 training roc_auc binary 0.599
2 testing roc_auc binary 0.439
$pmse
# A tibble: 2 × 4
.source .pmse .null_pmse .pmse_ratio
<fct> <dbl> <dbl> <dbl>
1 training 0.00798 0.00689 1.16
2 testing 0.00880 0.00691 1.27
$specks
# A tibble: 2 × 2
.source .specks
<fct> <dbl>
1 training 0.201
2 testing 0.155
attr(,"class")
[1] "discrimination"6.2 Additional Functionality
6.2.1 Grouping
Many utility metrics include a group_by argument to group the metrics by group during calculation. For example, this code calculates moments by species.
# A tibble: 60 × 7
species variable statistic original synthetic difference
<fct> <fct> <fct> <dbl> <dbl> <dbl>
1 Adelie bill_length_mm count 146 153 7
2 Chinstrap bill_length_mm count 68 78 10
3 Gentoo bill_length_mm count 119 102 -17
4 Adelie bill_length_mm mean 38.8 38.5 -0.294
5 Chinstrap bill_length_mm mean 48.8 47.3 -1.56
6 Gentoo bill_length_mm mean 47.6 48.0 0.475
7 Adelie bill_length_mm sd 2.66 2.93 0.267
8 Chinstrap bill_length_mm sd 3.34 3.07 -0.273
9 Gentoo bill_length_mm sd 3.11 3.40 0.299
10 Adelie bill_length_mm skewness 0.156 0.505 0.349
# ℹ 50 more rows
# ℹ 1 more variable: proportion_difference <dbl>6.2.2 Weighting
Many utility metrics include a weight_var argument to use weighted statistics during calculation. For example, this code weights the moments by the body weight of the penguins.
# A tibble: 20 × 6
variable statistic original synthetic difference proportion_difference
<fct> <fct> <dbl> <dbl> <dbl> <dbl>
1 bill_length_mm count 1.40e+6 1.39e+6 -1.5 e+4 -0.0107
2 bill_length_mm mean 4.46e+1 4.41e+1 -4.71e-1 -0.0106
3 bill_length_mm sd 5.38e+0 5.54e+0 1.60e-1 0.0297
4 bill_length_mm skewness -7.39e-2 -5.74e-2 1.64e-2 -0.222
5 bill_length_mm kurtosis -7.89e-1 -9.15e-1 -1.26e-1 0.160
6 bill_depth_mm count 1.40e+6 1.39e+6 -1.5 e+4 -0.0107
7 bill_depth_mm mean 1.70e+1 1.71e+1 1.28e-1 0.00754
8 bill_depth_mm sd 2.01e+0 1.94e+0 -7.07e-2 -0.0351
9 bill_depth_mm skewness 1.05e-2 -1.37e-1 -1.47e-1 -14.0
10 bill_depth_mm kurtosis -1.00e+0 -9.17e-1 8.70e-2 -0.0867
11 flipper_length… count 1.40e+6 1.39e+6 -1.5 e+4 -0.0107
12 flipper_length… mean 2.03e+2 2.01e+2 -1.97e+0 -0.00970
13 flipper_length… sd 1.44e+1 1.46e+1 1.60e-1 0.0111
14 flipper_length… skewness 1.48e-1 3.99e-1 2.51e-1 1.70
15 flipper_length… kurtosis -1.14e+0 -1.04e+0 1.08e-1 -0.0940
16 body_mass_g count 1.40e+6 1.39e+6 -1.5 e+4 -0.0107
17 body_mass_g mean 4.36e+3 4.31e+3 -5.19e+1 -0.0119
18 body_mass_g sd 8.26e+2 8.17e+2 -9.84e+0 -0.0119
19 body_mass_g skewness 2.69e-1 4.64e-1 1.95e-1 0.724
20 body_mass_g kurtosis -9.70e-1 -7.26e-1 2.44e-1 -0.251 Most commonly, weight_var is used when synthesizing data from surveys.