8 Example 01: Penguins
This example synthesizes the penguins data set demonstrated in the last few chapters.
8.1 Setup
The example uses complete observations and a subset of variables from the Palmer Penguins data set (cite!).
8.2 Start Data
The synthesis starts with a bootstrap sample of species, island, and sex. The number of observations synthesized will exactly equal the number of observations in the confidential data set.
8.3 Roadmap
We use correlation order with bill_length_mm as the reference variable.
8.4 Synth Spec
# use library(parsnip) and library(recipes) to create specifications for
# each variable
rpart_mod <- parsnip::decision_tree() %>%
parsnip::set_mode("regression") %>%
parsnip::set_engine("rpart")
synth_spec <- synth_spec(
roadmap = roadmap,
synth_algorithms = rpart_mod,
recipes = construct_recipes(roadmap = roadmap),
predict_methods = sample_rpart
)8.5 Extras
# don't add extra noise to predictions
noise <- noise(
roadmap = roadmap,
add_noise = FALSE,
exclusions = 0
)
# don't impose constraints
constraints <- constraints(
roadmap = roadmap,
constraints = NULL,
max_z = 0
)
# only generate one synthetic data set
replicates <- replicates(
replicates = 1,
workers = 1,
summary_function = NULL
)8.6 Synthesize
8.7 Evaluate
data_combined <- bind_rows(
"confidential" = penguins_complete,
"synthetic" = synth1$synthetic_data,
.id = "source"
)
data_combined %>%
select(source, bill_length_mm, bill_depth_mm, flipper_length_mm, body_mass_g) %>%
pivot_longer(-source, names_to = "variable") %>%
ggplot(aes(value, fill = source)) +
geom_density(alpha = 0.4, color = NA) +
facet_wrap(~variable, scales = "free") +
theme(
strip.background = element_blank(),
panel.grid = element_blank()
) +
guides(fill = "none")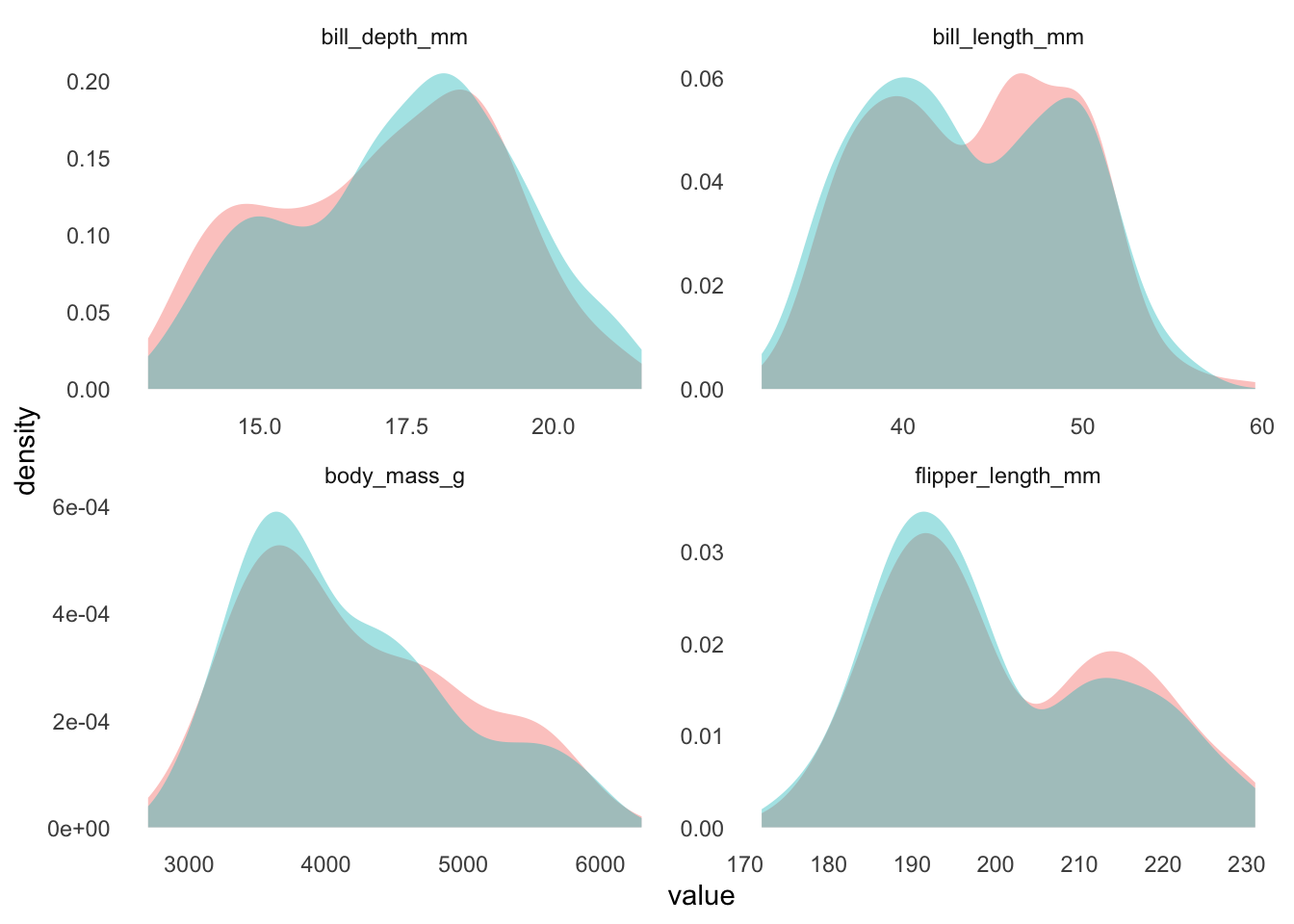
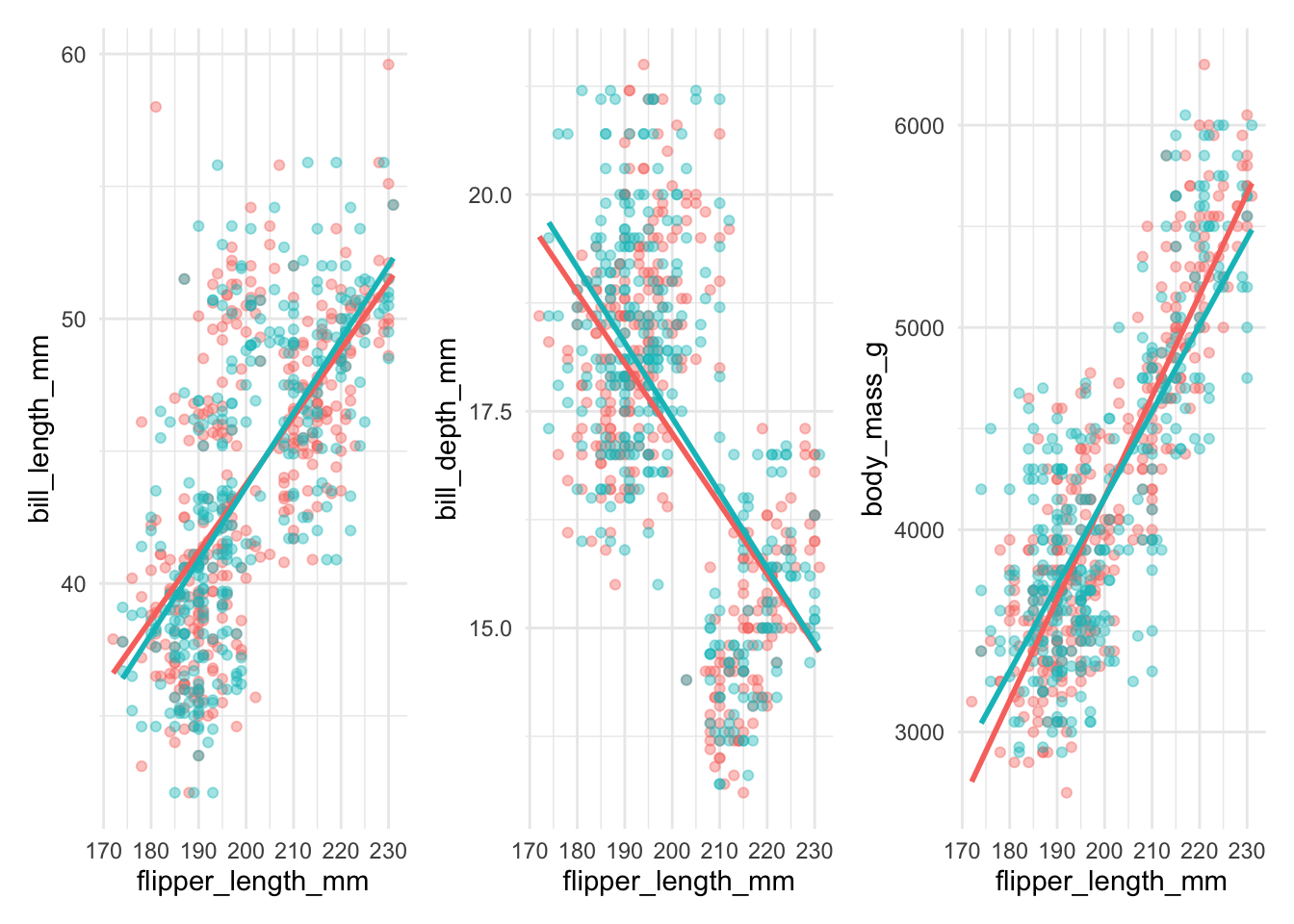
pane1 <- data_combined %>%
ggplot(aes(flipper_length_mm, bill_length_mm, color = source)) +
geom_point(alpha = 0.4) +
geom_smooth(method = "lm", se = FALSE) +
guides(color = "none")
pane2 <- data_combined %>%
ggplot(aes(flipper_length_mm, bill_depth_mm, color = source)) +
geom_point(alpha = 0.4) +
geom_smooth(method = "lm", se = FALSE) +
guides(color = "none")
pane3 <- data_combined %>%
ggplot(aes(flipper_length_mm, body_mass_g, color = source)) +
geom_point(alpha = 0.4) +
geom_smooth(method = "lm", se = FALSE) +
guides(color = "none")
pane1 + pane2 + pane3`geom_smooth()` using formula = 'y ~ x'
`geom_smooth()` using formula = 'y ~ x'
`geom_smooth()` using formula = 'y ~ x'