9 Example 02: Diamonds
9.1 Setup
9.1.1 The data
We synthesize a subset of the diamonds data set to demonstrate creating multiple syntheses with tidysynthesis.
9.2 Synthesis
9.2.1 Start data
We start with a joint simple random sample of cut and clarity. We synthesize the same number of observations as the original data.
9.2.2 Visit sequence
carat seems like an important variable based on exploratory data analysis and some subject matter knowledge. We synthesize in order from most to least correlated with carat.
9.2.3 roadmap
The roadmap combines the confidential data, starting data, and visit sequence. This object will inform the remaining synthesis objects.
9.2.4 synthspec
We use regression trees with node sampling and no feature/target engineering to synthesize the data.
9.2.5 noise
For the first synthesis, we don’t add extra noise to predicted values.
9.2.6 constraints
We don’t use constraints or replicates for this synthesis.
9.2.7 presynth
9.2.8 synthesize
9.3 Evaluation
We use several different metrics to evaluate the synthetic data.
9.3.1 Univariate
First, we look at summary statistics with util_moments() from library(syntheval). The synthetic data do a great job of recreating most summary statistics from the original data.
summary_stats <- util_moments(
synth1,
diamonds_prepped
)
summary_stats %>%
filter(statistic == "mean")# A tibble: 7 × 6
variable statistic original synthetic difference proportion_difference
<fct> <fct> <dbl> <dbl> <dbl> <dbl>
1 carat mean 0.658 0.658 -0.000204 -0.000310
2 x mean 5.42 5.42 0.00579 0.00107
3 y mean 5.42 5.42 -0.00103 -0.000190
4 z mean 3.34 3.34 -0.000537 -0.000161
5 price mean 3170. 3170. 0.0303 0.00000955
6 table mean 57.4 57.4 -0.0192 -0.000335
7 depth mean 61.7 61.7 0.0181 0.000294 # A tibble: 7 × 6
variable statistic original synthetic difference proportion_difference
<fct> <fct> <dbl> <dbl> <dbl> <dbl>
1 carat sd 0.360 0.358 -0.00113 -0.00313
2 x sd 0.939 0.937 -0.00198 -0.00211
3 y sd 0.936 0.935 -0.000607 -0.000648
4 z sd 0.579 0.573 -0.00519 -0.00898
5 price sd 3357. 3324. -32.4 -0.00966
6 table sd 2.21 2.18 -0.0287 -0.0130
7 depth sd 1.41 1.44 0.0319 0.0226 # A tibble: 7 × 6
variable statistic original synthetic difference proportion_difference
<fct> <fct> <dbl> <dbl> <dbl> <dbl>
1 carat skewness 1.29 1.28 -0.0106 -0.00822
2 x skewness 0.558 0.568 0.00950 0.0170
3 y skewness 0.549 0.524 -0.0253 -0.0460
4 z skewness 0.550 0.538 -0.0120 -0.0219
5 price skewness 2.11 2.07 -0.0361 -0.0172
6 table skewness 0.747 0.711 -0.0364 -0.0487
7 depth skewness -0.258 -0.158 0.0996 -0.387 # A tibble: 7 × 6
variable statistic original synthetic difference proportion_difference
<fct> <fct> <dbl> <dbl> <dbl> <dbl>
1 carat kurtosis 2.04 1.98 -0.0561 -0.0275
2 x kurtosis -0.257 -0.228 0.0286 -0.112
3 y kurtosis -0.282 -0.363 -0.0811 0.288
4 z kurtosis -0.261 -0.535 -0.274 1.05
5 price kurtosis 4.67 4.52 -0.146 -0.0313
6 table kurtosis 1.09 0.744 -0.348 -0.319
7 depth kurtosis 2.56 3.27 0.709 0.277 We also look at important percentiles.
eval_data <- bind_rows(
"confidential" = diamonds_prepped,
"synthetic" = synth1$synthetic_data,
.id = "source"
)
eval_data %>%
select(source, where(is.numeric)) %>%
group_by(source) %>%
summarize(
quantile = seq(0.05, 0.95, 0.05),
across(everything(), .fns = ~quantile(.x, probs = seq(0.05, 0.95, 0.05)))
) %>%
ungroup() %>%
pivot_longer(
c(-source, -quantile),
names_to = "variable",
values_to = "value"
) %>%
pivot_wider(names_from = source, values_from = value) %>%
ggplot(aes(confidential, synthetic)) +
geom_abline(intercept = 0, slope = 1) +
geom_point(alpha = 0.1) +
#coord_equal() +
facet_wrap(~ variable, scales = "free")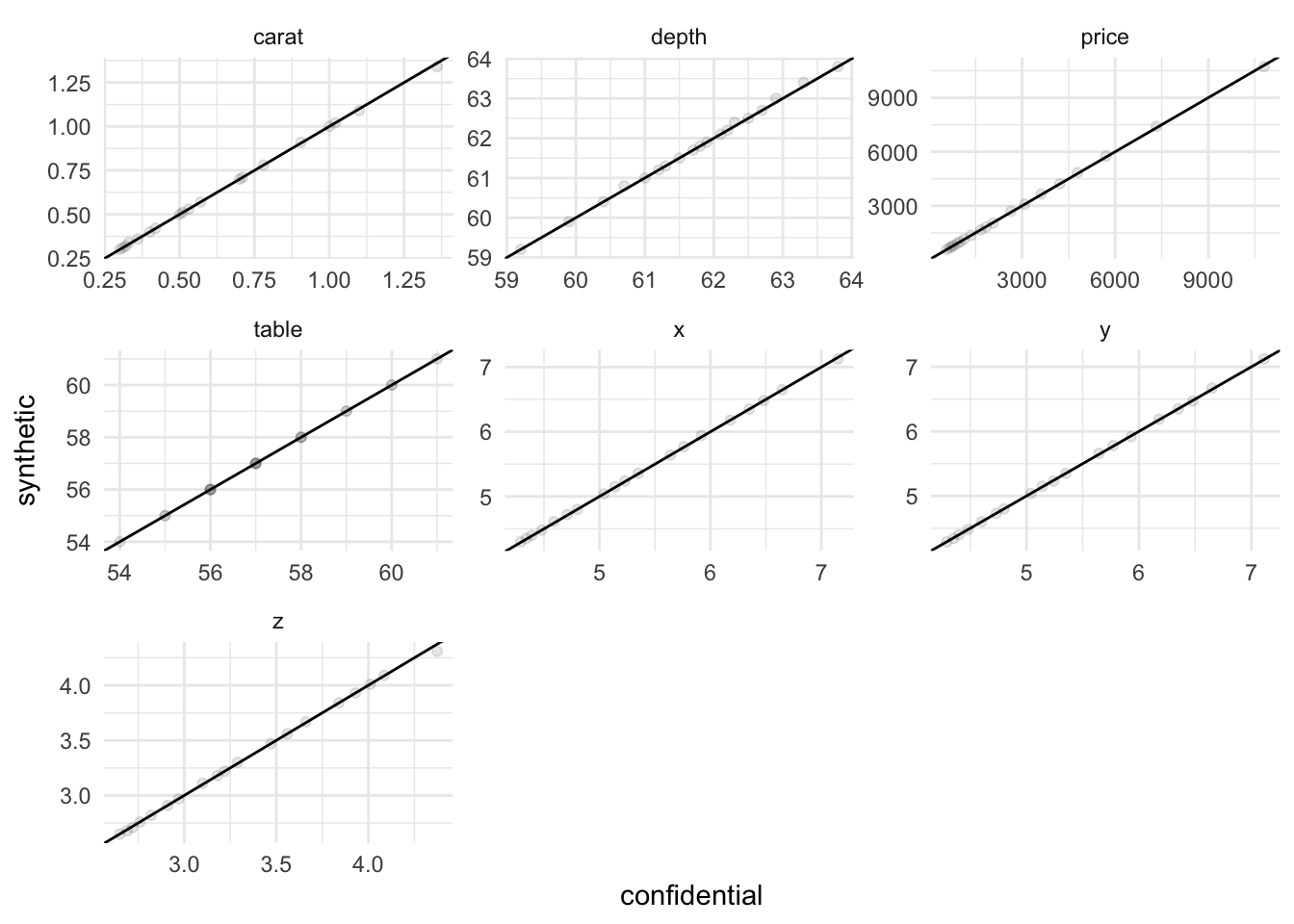
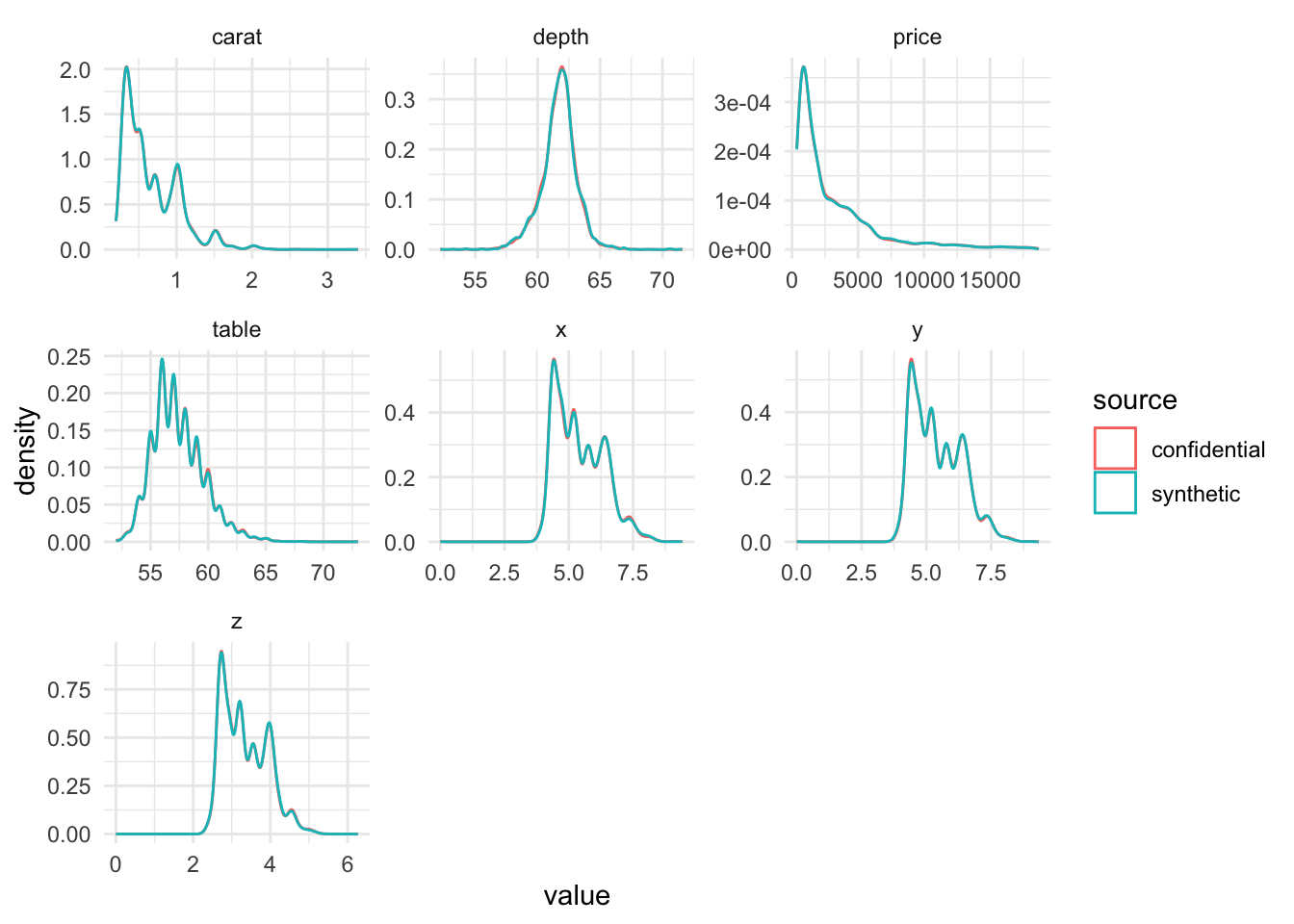
We look at density plots. The original data and synthetic data have comparable distributions for all variables.
9.3.2 Multivariate
We look at a few key relationships. It is difficult to distinguish between the original data and synthetic data.
eval_data %>%
ggplot(aes(carat, price, color = source)) +
geom_point(alpha = 0.05) +
geom_smooth(method = "lm")`geom_smooth()` using formula = 'y ~ x'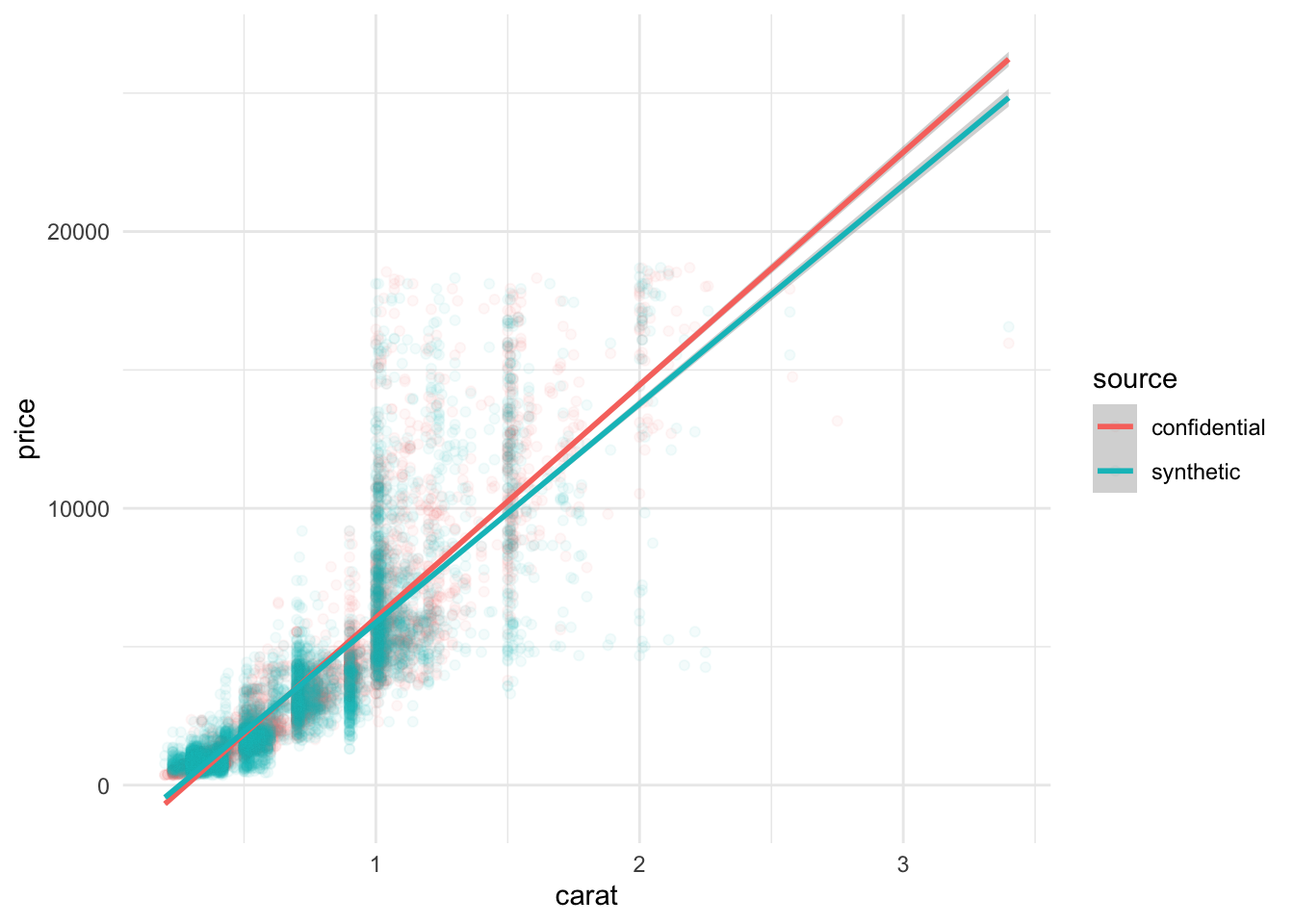
We look at how well the synthetic data recreate the correlations in the original data.
This means that the average correlation difference is off by about 0.05 in the synthetic data.
carat x y z price table depth
carat NA NA NA NA NA NA NA
x -0.04 NA NA NA NA NA NA
y -0.05 -0.03 NA NA NA NA NA
z -0.04 -0.03 -0.04 NA NA NA NA
price -0.05 -0.05 -0.06 -0.04 NA NA NA
table -0.05 -0.04 -0.04 0.00 -0.01 NA NA
depth 0.01 0.06 0.05 -0.06 0.02 0.19 NAThe errors are pretty even across the correlation matrix.
disc1 <- discrimination(postsynth = synth1, data = diamonds_prepped)
rpart_rec <- recipe(
.source_label ~ .,
data = disc1$combined_data
)
rpart_mod <- decision_tree(cost_complexity = 0.001) %>%
set_mode(mode = "classification") %>%
set_engine(engine = "rpart")
disc1 <- disc1 %>%
add_propensities(
recipe = rpart_rec,
spec = rpart_mod
)
disc1 <- disc1 %>%
add_pmse %>%
add_pmse_ratio(times = 100)
disc1$pmse# A tibble: 2 × 4
.source .pmse .null_pmse .pmse_ratio
<fct> <dbl> <dbl> <dbl>
1 training 0.140 0.0115 12.3
2 testing 0.141 0.0114 12.3The pMSE ratio is a discriminant-based metric. The value of the pMSE ratio is good but not great in this case.
9.4 Approach #2
We create a second synthesis. This time, we add extra noise to predicted values when synthesizing the data. Fortunately, we can reuse many of the objects created in the first synthesis. Here, we update the noise object and the presynth object. Then we synthesize again.
We add extra noise to our predicted values. We use 400 ntiles, which works out to about 17 observations per ntile.
The original data has limited precision. We round the synthetic variables to match this precision.
synth2$synthetic_data$table <- round(synth2$synthetic_data$table, digits = 0)
synth2$synthetic_data$carat <- round(synth2$synthetic_data$carat, digits = 2)
synth2$synthetic_data$y <- round(synth2$synthetic_data$y, digits = 2)
synth2$synthetic_data$x <- round(synth2$synthetic_data$x, digits = 2)
synth2$synthetic_data$z <- round(synth2$synthetic_data$z, digits = 2)
synth2$synthetic_data$price <- round(synth2$synthetic_data$price, digits = 0)
synth2$synthetic_data$depth <- round(synth2$synthetic_data$depth, digits = 1)Most of the utility metrics look comparable to the first synthesis; however, the pMSE ratio increases modestly.
disc2 <- discrimination(postsynth = synth2, data = diamonds_prepped)
rpart_rec <- recipe(
.source_label ~ .,
data = disc1$combined_data
)
rpart_mod <- decision_tree(cost_complexity = 0.001) %>%
set_mode(mode = "classification") %>%
set_engine(engine = "rpart")
disc2 <- disc2 %>%
add_propensities(
recipe = rpart_rec,
spec = rpart_mod
)
disc2 <- disc2 %>%
add_pmse %>%
add_pmse_ratio(times = 100)
disc2$pmse# A tibble: 2 × 4
.source .pmse .null_pmse .pmse_ratio
<fct> <dbl> <dbl> <dbl>
1 training 0.148 0.0116 12.7
2 testing 0.147 0.0116 12.6- add hyperparameters
- add evaluation
- add constraints